INTRODUCTION
The challenge of keeping pace with the exponential growth in the scientific literature is a significant barrier to effectively translating research evidence into practice. This growing gap between knowledge and application often leads to overlooking valuable evidence, emphasizing the need for efficient strategies to navigate and integrate this vast amount of information. 1 Addressing the replication crisis and improving scientific research practices have become crucial in this context. Strategies such as pre-registration of research, peer reviews throughout the research process, and promoting open science practices are vital for maintaining scientific integrity and trust. 2
Moreover, the recent global pandemic has underscored the challenges in quality control and the need for reliable information amidst a surge in biomedical publications, exacerbating the difficulty of keeping up-to-date with the latest scientific findings. 3 The pollution of scientific literature, or “infollution,” necessitates a critical evaluation of current peer-review systems and strategies to enhance the reproducibility of published data, aiming to curtail the dissemination of misleading or low-quality research. 4 Furthermore, the pressure to publish for career advancement can lead to a prioritization of quantity over quality, undermining the validity of scientific research. As an incentive, this publication strategy is detrimental to conducting high-quality scientific research and, thus, also prevents advances in health services. 5 Strategies for handling knowledge gaps and conducting systematic reviews are crucial for navigating these challenges and fostering a culture prioritizing scientific integrity and relevance. 6 Expert reviews can offer researchers a starting point when entering a new domain. However, such reviews are typically outdated quickly in fast-moving research fields. To remain succinct and accessible, expert reviews give only a high-level view and often focus on specific questions and topics. Interested researchers, thus, have to engage in systematic and time-consuming literature reviews, in which it is challenging to recognize high-quality research.
Large language models (LLMs) are revolutionizing how scientists access and utilize the vast repository of scientific knowledge. LLMs can dramatically improve their response accuracy by integrating tools like retrieval-augmented generation (RAG) from the scientific literature. This technique enables the dynamic incorporation of information from external databases, such as PubMed or Google Scholar, ensuring that the provided information is factual and up-to-date. As outlined by Lewis et al. 7 in their seminal work on RAG for knowledge-intensive Natural Language Processing (NLP) tasks, this approach combines the extensive database of algorithms with an intuitive, natural chatting manner, offering a seamless and efficient way for scientists to retrieve relevant scientific literature. By building a RAG tool for semantic similarity-based literature retrieval, LLMs can provide contextual user assistance, effectively bridging the gap between complex scientific queries and the vast expanse of existing scientific research. This integration not only empowers scientists with the fastest access to information in a conversational manner but also leverages factual information to answer questions based on scientific articles, thereby facilitating a more informed and evidence-based scientific inquiry. Importantly, the effectiveness of RAG hinges on the quality and relevance of the curated literature collection used as its knowledge base, emphasizing the need for expert-curated databases to ensure accurate and reliable information retrieval.
We previously performed a community-driven literature search and evaluation of drug repurposing tools, reviews, and databases as part of the REPO4EU consortium (https://repo4.eu/). 8 Our article curation method, detailed in the Methods section, ensured a rigorous selection of high-quality, relevant, and publicly available publications in the drug repurposing field. This carefully curated collection serves as the basis for a new tool we created to help researchers overcome the difficulties they encounter when sifting through the enormous and disorganized amount of drug repurposing data.
RESULTS
To address the pressing need for efficient access to high-quality drug repurposing information, we developed “DrugRepoChatter,” an LLM-powered chatbot designed to serve as a fundamental resource for academics, scientists, and drug repurposing enthusiasts. DrugRepoChatter aims to combine the fragmented landscape of drug repurposing literature into a cohesive and accessible format. This tool utilizes our curated collection of scientific literature as its knowledge base. DrugRepoChatter employs RAG technology to dynamically incorporate information from this curated list, enabling immediate, accurate, and contextually relevant interactions. By simplifying access to valuable scientific information, DrugRepoChatter aims to accelerate drug repurposing research and facilitate more informed decision-making in the field.
DrugRepoChatter is an advancement toward democratizing access to scientific knowledge in drug repurposing, making a wealth of expert-curated open-access literature accessible to a wide audience. DrugRepoChatter is the direct result of the REPO4EU consortium’s efforts that aim to foster a more involved and knowledgeable community on drug repurposing, hence expediting the discovery and use of repurposed drugs.
Figure 1 illustrates the process and architecture of DrugRepoChatter, showing how the chatbot processes and retrieves information from the curated articles. DrugRepoChatter uses an LLM model to embed text splits from expert-selected open-access articles into a vector space for similarity searches. This system enables the chatbot to match user queries with relevant literature effectively, providing clear and accurate responses based on the embedded scientific content.
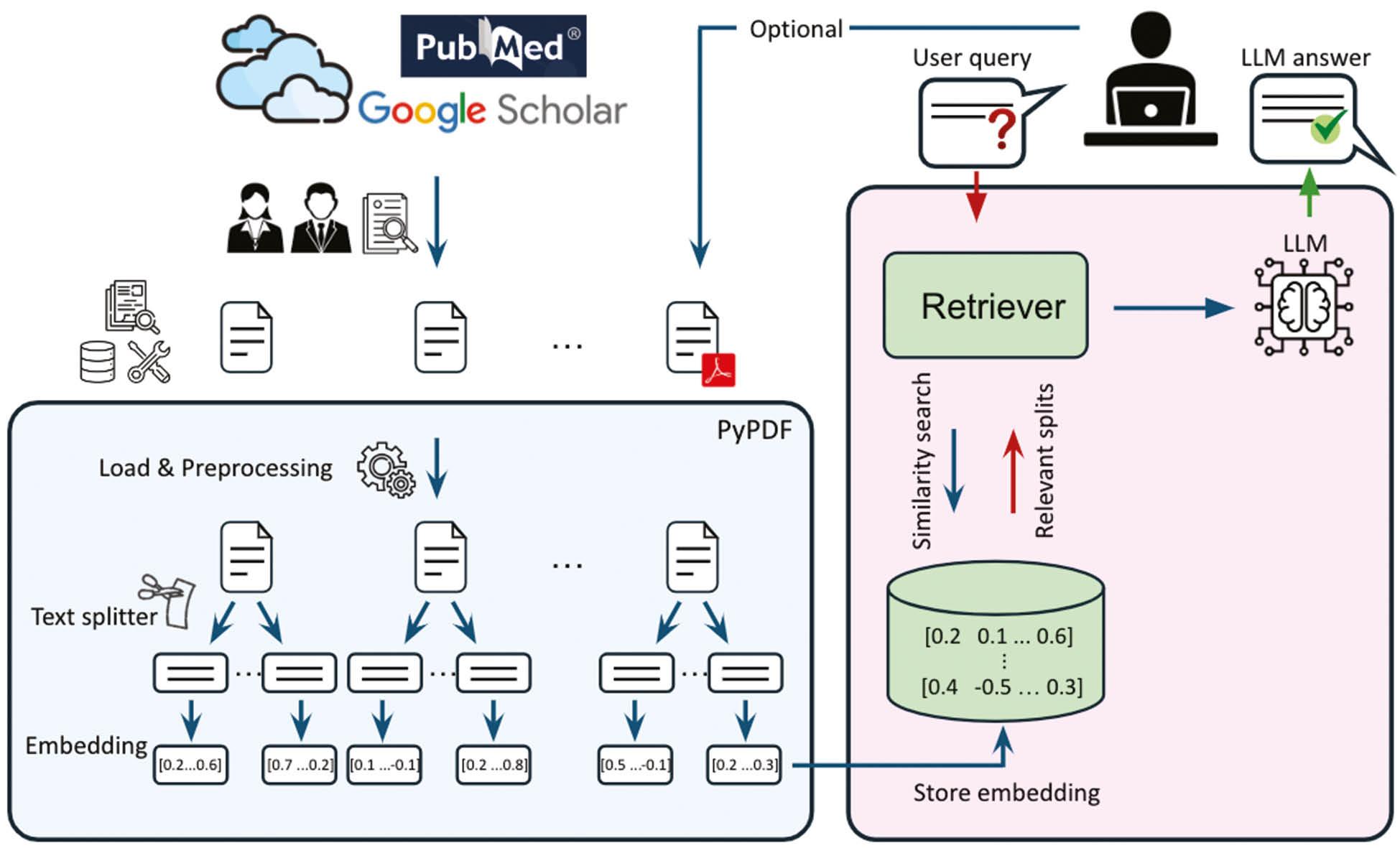
A schematic representation of DrugRepoChatter. Initially, 285 drug repurposing articles have been carefully selected by experts. The articles, in PDF format, have been loaded, preprocessed, and split into chunks. Each text split has been embedded into a vector space using an LLM model and stored. These data are then used for similarity searches, where the embedding of the user query is compared to the embedding of text splits. Then, the most relevant splits are selected by the retriever. Next, the relevant splits are used by the LLM model to generate answers. Users can further personalize the database by adding their own papers. LLM, large language model.
DrugRepoChatter is based on a database containing 285 expert-curated open-access articles selected for their relevance and quality in drug repurposing. This curation process was critical in ensuring that the chatbot could provide accurate, informed responses to user queries, significantly enhancing the efficiency of literature review processes for researchers. Following the open science paradigm, we ensured unrestricted access to this information by focusing solely on open-access publications.
DrugRepoChatter’s design and technology enable it to match user queries with relevant articles from the database effectively. Users receive answers informed by the embedded literature, presented clearly and accessibly, streamlining the process of obtaining scientific information. The tool can help integrate scattered information into a cohesive, user-friendly format, eliminating the need for researchers to manually navigate multiple databases or sift through extensive volumes of literature.
The acceleration of discovery in drug repurposing is another notable outcome of DrugRepoChatter’s implementation. By providing quick access to the scientific literature and equipping researchers with the necessary tools and information, DrugRepoChatter supports identifying novel drug repurposing opportunities and increases the pace of discovery in this field.
Feedback from initial users indicates a strong appreciation for the chatbot’s intuitive design and the credibility of its responses, underscoring the value of direct source citation. Overall, DrugRepoChatter exemplifies the transformative potential of leveraging advanced technologies to support scientific inquiry. Its development and successful implementation demonstrate a significant advancement in how researchers access and interact with scientific literature, fostering a more efficient and informed exploration of new therapeutic opportunities.
DISCUSSION
The development and implementation of DrugRepoChatter represent a significant advancement in how researchers access and interact with scientific literature. By leveraging advanced technologies such as vector databases, semantic similarity searches, and RAG, our chatbot provides accurate, informed responses to user queries, significantly enhancing the efficiency of the literature review processes for researchers.
The careful curation of scientific articles was critical in ensuring that DrugRepoChatter could provide reliable, informed answers. By focusing exclusively on open-access publications, we promote an open science approach and ensure unrestricted access to this information. This curated database is the foundation of DrugRepoChatter’s ability to match user queries with relevant articles effectively, providing users with clear, accessible answers that streamline the process of accessing scientific information.
DrugRepoChatter eliminates the need for researchers to manually navigate multiple databases or sift through extensive volumes of literature. Its design and technology enable it to accelerate education in drug repurposing by providing quick access to the relevant scientific literature and equipping researchers with the necessary tools and information. This positive reception highlights the tool’s potential to foster more efficient and informed exploration of new therapeutic opportunities by researchers. The interaction data collected from these users will be instrumental in further refining DrugRepoChatter, ensuring it continues to serve the research community’s needs effectively.
CONCLUSION AND FUTURE RESEARCH
In conclusion, DrugRepoChatter exemplifies the transformative potential of leveraging advanced technologies to support scientific inquiry. Its development and successful implementation demonstrate that researchers can access and interact with the scientific literature more efficiently and informedly, ultimately fostering a more productive exploration of new therapeutic opportunities in drug repurposing.
DrugRepoChatter facilitates a more streamlined review process, enabling researchers to stay abreast of the latest developments in their field. The chatbot’s positive reception by the research community highlights its potential to significantly impact how scientific literature is accessed and utilized in drug repurposing research.
Future research will focus on expanding and updating the chatbot’s database through a systematic and efficient process. We have already created targeted PubMed queries for drug repurposing reviews, databases, and tools as part of the REPO4EU community effort. We intend to use these customized queries to create a periodic querying system that consistently pulls new publications from PubMed. Basic filtering criteria, like choosing only articles with valid PubMed IDs, will be applied during this process. We will incorporate the new articles’ titles and abstracts into our knowledge base to produce time-stamped versions. This approach allows us to continually expand our knowledge base without the computational demands and potential user experience issues that could arise from embedding and chatting with full-text articles, even if limited to open-access publications. By using this approach, DrugRepoChatter will be able to remain up to date with the most recent research while still being user-friendly and efficient. The process of updating articles will be streamlined because they will not need to be re-embedded into the database. Users will always have access to the most recent information without overloading the system or degrading response times thanks to this approach, which strikes a compromise between comprehensiveness and practicality.
Additionally, further development will aim to enhance the chatbot’s natural language processing capabilities in order to improve the relevance and accuracy of its responses. As new open-source LLMs are released, we plan to potentially replace the currently running model with newer, more advanced versions. This ongoing update of the underlying LLM could significantly benefit future users by providing improved performance, expanded capabilities, and potentially better alignment with the specific needs of drug repurposing research.
As artificial intelligence (AI) and machine learning continue to advance, there is significant potential for tools like DrugRepoChatter to become integral components of the research ecosystem, facilitating faster and more informed scientific inquiry. Continued collaboration between AI developers and the scientific community will be crucial in realizing this potential and ensuring that such tools are tailored to meet the evolving needs of researchers.
METHODS
The REPO4EU consortium aims to develop a platform that sets global standards for drug repurposing, both in clinical and information management aspects. Partners conducted a literature review and gathered scientific papers on drug repurposing to create a knowledge base for this platform.
Article Curation Process
The initial phase involved compiling an exhaustive list of potential articles from databases such as PubMed 9 and Google Scholar. 10 This list was refined through a meticulous review process that assessed each article’s relevance, quality, and contribution to drug repurposing. Our article curation strategy began with identifying relevant and novel research papers, reviews, and databases in the drug repurposing field. Following this first step, we conducted a thorough evaluation to assess the quality and relevance of the literature in light of current research trends and clinical utility. In particular, we applied the following inclusion criteria to filter the publications for our study:
Tools and methods have to be usable in a programmatic fashion by including the terms package, Application Programming Interface (API), or code. Regarding tools designed for users who are not usually expected to have programming knowledge (e.g., clinicians), a web app or Graphical User Interface (GUI) should be provided.
Databases and data resources should contain the term database.
We consider publications to be accepted by the community if they have more than 30 PubMed citations. Publications with fewer citations are only considered if published within the last 5 years.
All publications should be open-access.
This approach guaranteed that only publications that met our high scientific validity requirements and were publicly available made it into our refined selection. Special attention was given to ensuring that only open-access articles were included to facilitate unrestricted access. The selected articles were cataloged based on their relevance to different categories within drug repurposing, such as databases, reviews, and tools. This organization facilitated easier access and retrieval of information relevant to specific queries or topics. For each article, we obtained its corresponding PDF version.
Embedding Articles in the DrugRepoChatter Vector Database
To extract text from the curated articles’ PDF files, we utilized the PyPDFLoader module from the LangChain-community Python package. 11 We configured PyPDFLoader to extract text from each page of the PDF as a separate document while storing the page numbers in the metadata. This involved extracting the text while discarding references to focus on the core content. Additional article information, like title, abstract, and DOI, was included as metadata.
The text extracted from the articles was then embedded using the nomic-embed-text model. 12 These embeddings were saved in a Facebook AI Similarity Search (FAISS) vector store, 13 a library developed by Facebook for efficient similarity searches. FAISS uses clustering methods, such as k-means, to organize the feature space, allowing for the quick identification of the cluster nearest to a queried vector. This facilitates efficient and rapid text retrieval based on the similarity and relevance of user queries. This functionality is essential for DrugRepoChatter’s ability to quickly source and cite pertinent articles in response to user inquiries.
Chatbot Implementation
We built DrugRepoChatter using LangChain 11 as our foundation, a powerful library for constructing LLM applications with RAG capabilities. This technique allows the LLM to dynamically tap into external knowledge sources, like our expert-curated database, to deliver accurate and up-to-date information in its responses. 7,14,15
To make DrugRepoChatter user-friendly, we incorporated the Llama3:8b model. 16 This system allows for fine-tuning of search results through several key parameters: score_threshold, k, and fetch-k. These parameters can be adjusted via the web interface, giving researchers control over the retrieval process. The score threshold, defaulting to 0.9, can be adjusted from 0.0 to 1.0 with a step size of 0.1, allowing users to set the minimum relevance score for retrieved documents. The k parameter, which determines the number of results returned, defaults to 3 and can be modified between 1 and 50. Similarly, the fetch-k parameter, controlling the initial retrieval scope before filtering, defaults to 20 and is also adjustable from 1 to 50. These customization options ensure that the chatbot’s answers are tailored to the user’s specific query needs. While the model’s temperature is set to 0.0 by default in the ollama_connector script to minimize hallucinations, this is not directly controlled by users in the GUI. LangChain plays a crucial role in seamlessly bridging the gap between the chatbot interface and the Llama3 models, ultimately empowering DrugRepoChatter to deliver precise and insightful responses based on the curated literature.
DrugRepoChatter retrieves the most relevant articles’ embeddings to generate informed and accurate responses when posed with queries. This system ensures that the chatbot’s answers are only grounded in the embedded scientific literature. To provide responses that are concise and easy to understand, DrugRepoChatter was designed to adapt its answers based on the retrieved articles. It can also admit when it does not have enough information, ensuring that users are provided with informative and trustworthy responses.
We used Streamlit 17 to implement our chatbot, a faster way to build and share data apps. Streamlit turns data scripts into shareable web apps. This allows us to create a user-friendly interface for DrugRepoChatter, making it accessible and easy-to-use for researchers and other users.


